Analyzing the mesh#
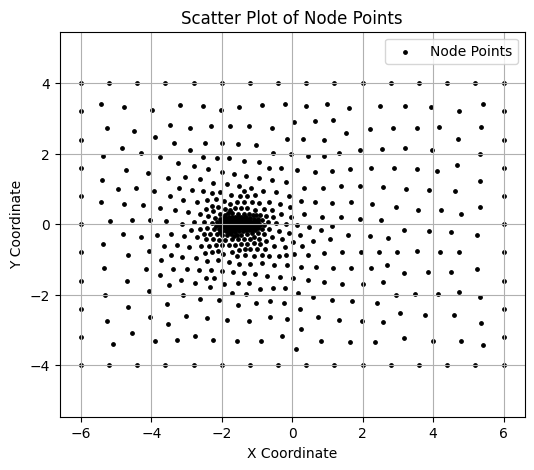
Visualizing the node points.#
The node.npy file is loaded and visualized. It contains a list of (x,y) coordinate values.
# prompt: Write code to load a file from google drive
import jax.numpy as jnp
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
node = jnp.load('fig/airfoil0.node.npy')
# Create a scatter plot of the node points
plt.figure(figsize=(6, 5))
plt.scatter(node[:,0], node[:,1], color='black', marker='o', s=6, label="Node Points") # Customize color and size as needed
plt.xlabel("X Coordinate")
plt.ylabel("Y Coordinate")
plt.title("Scatter Plot of Node Points")
plt.legend()
plt.grid(True)
plt.axis('equal') # Ensures the plot scales equally in both directions
(np.float64(-6.6), np.float64(6.6), np.float64(-4.4), np.float64(4.4))
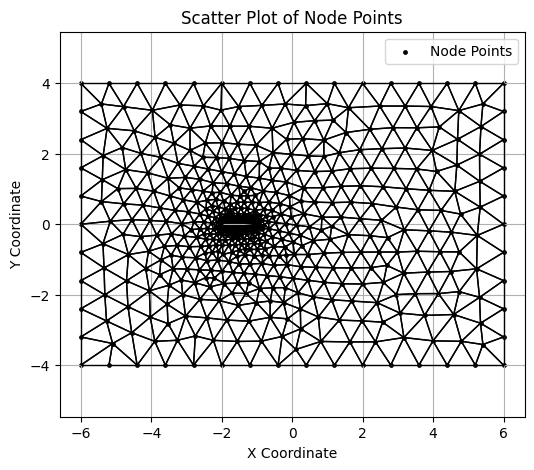
Visualizing the mesh#
elem.npy file has the information on the nodes that form each of the cell triangle.
elem = jnp.load('fig/airfoil0.elem.npy')
# Prepare line segments for each triangle
triangle_segments = []
centroids = []
for tri in elem:
# Get the nodes in the triangle
p1, p2, p3 = node[tri[0]], node[tri[1]], node[tri[2]]
# Define the lines for the triangle
triangle_segments.append([p1, p2])
triangle_segments.append([p2, p3])
triangle_segments.append([p3, p1])
centroids.append(jnp.mean(node[tri], axis=0))
# Create the scatter plot
plt.figure(figsize=(6, 5))
plt.scatter(node[:,0], node[:,1], color='black', marker='o', s=6, label="Node Points") # Customize color and size as needed
plt.xlabel("X Coordinate")
plt.ylabel("Y Coordinate")
plt.title("Scatter Plot of Node Points")
plt.legend()
plt.grid(True)
plt.axis('equal') # Ensures the plot scales equally in both directions
# Add triangle edges as a LineCollection
lc = LineCollection(triangle_segments, colors='black', linewidths=1)
plt.gca().add_collection(lc)
plt.show()
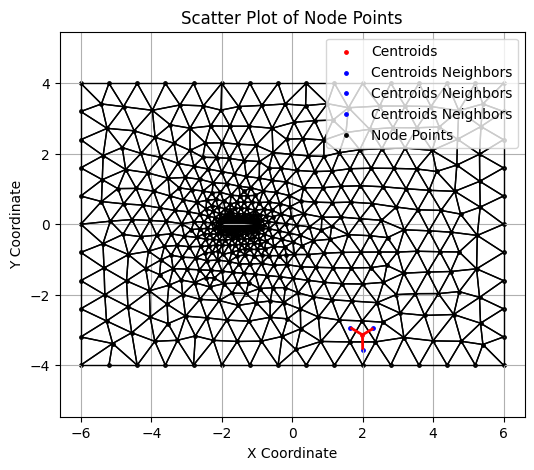
Visualizing the Neighbor information#
An example cell :
connect = jnp.load('fig/airfoil0.connect.npy')
# Plot centroids
i = 80
plt.figure(figsize=(6, 5))
plt.scatter(centroids[i][0], centroids[i][1], color='red', s=6, label="Centroids")
p0 = centroids[i]
neighbor_segments = []
for j in range(3):
p1 = centroids[connect[i][j]]
plt.scatter(p1[0], p1[1], color='blue', s=6, label="Centroids Neighbors")
neighbor_segments.append([p0, p1])
# Create the scatter plot
plt.scatter(node[:,0], node[:,1], color='black', marker='o', s=6, label="Node Points") # Customize color and size as needed
plt.xlabel("X Coordinate")
plt.ylabel("Y Coordinate")
plt.title("Scatter Plot of Node Points")
plt.legend()
plt.grid(True)
plt.axis('equal') # Ensures the plot scales equally in both directions
# Add triangle edges as a LineCollection
lc = LineCollection(triangle_segments, colors='black', linewidths=1)
plt.gca().add_collection(lc)
# Add triangle edges as a LineCollection
lc = LineCollection(neighbor_segments, colors='red', linewidths=2)
plt.gca().add_collection(lc)
plt.legend()
plt.grid(True)
plt.axis('equal')
plt.show()
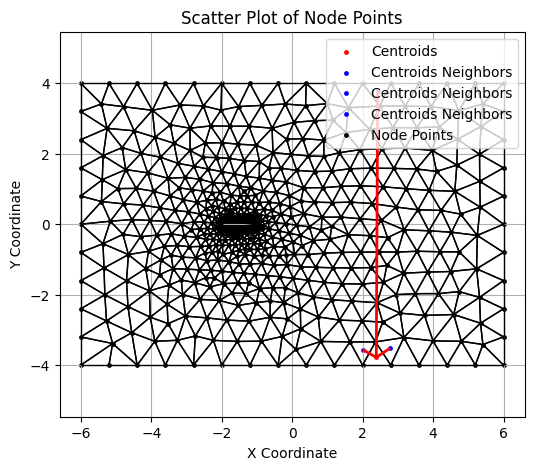
An example of cell with periodic boundary condition.
# Boundary cell
i = 315
plt.figure(figsize=(6, 5))
plt.scatter(centroids[i][0], centroids[i][1], color='red', s=6, label="Centroids")
p0 = centroids[i]
neighbor_segments = []
for j in range(3):
p1 = centroids[connect[i][j]]
plt.scatter(p1[0], p1[1], color='blue', s=6, label="Centroids Neighbors")
neighbor_segments.append([p0, p1])
# Create the scatter plot
plt.scatter(node[:,0], node[:,1], color='black', marker='o', s=6, label="Node Points") # Customize color and size as needed
plt.xlabel("X Coordinate")
plt.ylabel("Y Coordinate")
plt.title("Scatter Plot of Node Points")
plt.legend()
plt.grid(True)
plt.axis('equal') # Ensures the plot scales equally in both directions
# Add triangle edges as a LineCollection
lc = LineCollection(triangle_segments, colors='black', linewidths=1)
plt.gca().add_collection(lc)
# Add triangle edges as a LineCollection
lc = LineCollection(neighbor_segments, colors='red', linewidths=2)
plt.gca().add_collection(lc)
plt.legend()
plt.grid(True)
plt.axis('equal')
plt.show()
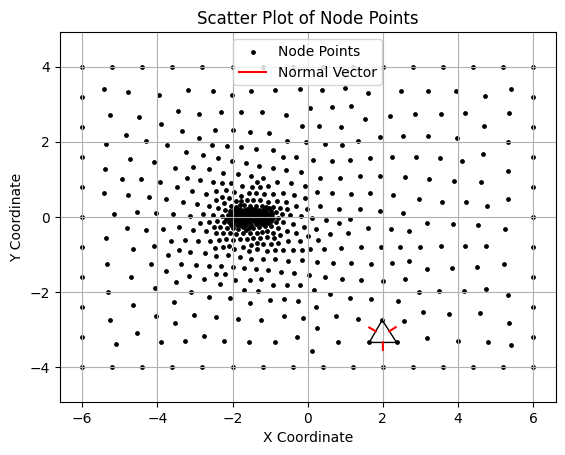
triangle_segments = []
normal_vectors = []
i = 80
p1, p2, p3 = node[elem[i][0]], node[elem[i][1]], node[elem[i][2]]
triangle_segments.extend([[p1, p2], [p2, p3], [p3, p1]])
# Calculate midpoints and normal vectors for each edge
edges = [(p1, p2), (p2, p3), (p3, p1)]
for edge in edges:
# Midpoint of the edge
mid_x = (edge[0][0] + edge[1][0]) / 2
mid_y = (edge[0][1] + edge[1][1]) / 2
# Vector along the edge
dx = edge[0][0] - edge[1][0]
dy = edge[0][1] - edge[1][1]
# Calculate the normal vector (perpendicular to the edge)
norm = jnp.sqrt(dx**2 + dy**2)
nx, ny = -dy / norm, dx / norm # Normalized perpendicular vector
# Scale the normal vector for plotting
normal_length = 0.2
normal_vectors.append(((mid_x, mid_y), (mid_x + normal_length * nx, mid_y + normal_length * ny)))
# Create the scatter plot
plt.scatter(node[:,0], node[:,1], color='black', marker='o', s=6, label="Node Points") # Customize color and size as needed
plt.xlabel("X Coordinate")
plt.ylabel("Y Coordinate")
plt.title("Scatter Plot of Node Points")
plt.legend()
plt.grid(True)
plt.axis('equal') # Ensures the plot scales equally in both directions
# Add triangle edges as a LineCollection
lc = LineCollection(triangle_segments, colors='black', linewidths=1)
plt.gca().add_collection(lc)
# Plot normal vectors at each edge's midpoint
for start, end in normal_vectors:
plt.plot([start[0], end[0]], [start[1], end[1]], 'r-', lw=1.5, label="Normal Vector" if 'Normal Vector' not in plt.gca().get_legend_handles_labels()[1] else None)
plt.legend()
plt.grid(True)
plt.axis('equal')
plt.show()